5.2 Group visualisation
library(tidyverse)
library(ggupset)
mat <- read_delim('DE_genes.txt.gz', delim='\t') %>% filter(group!='None') %>% arrange(time,desc(group)) %>% mutate(timegroup=str_c(time,'_',group)) %>% select(mgi_symbol, timegroup, logFC) %>% pivot_wider(names_from=timegroup, values_from=logFC, values_fill=list(logFC=0)) %>% column_to_rownames('mgi_symbol')
mat[mat!=0] <- 1
gp <- Upset(mat)
gp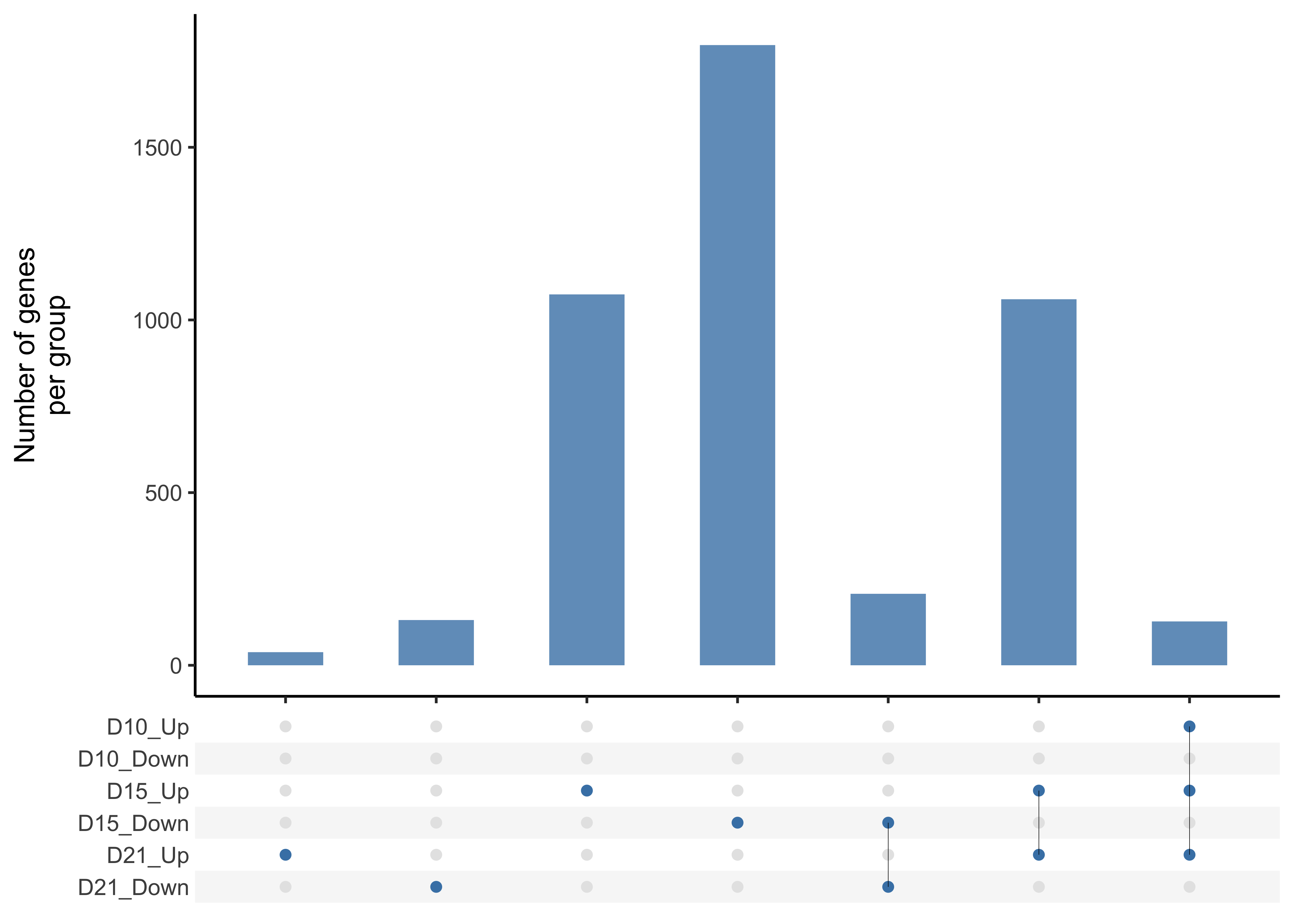
Also, extract members per gene group stored in df_full. The same as mat above but with 4 columns added ('code', 'ncode', 'freq' and 'member').
df_full <- gp$full %>% filter(freq>=30)
df_full
## # A tibble: 4,433 x 10
## code D10_Up D10_Down D15_Up D15_Down D21_Up D21_Down ncode freq member
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <chr>
## 1 1-0-1-0-1… 1 0 1 0 1 0 3 127 Il1a
## 2 1-0-1-0-1… 1 0 1 0 1 0 3 127 Gm4841
## 3 1-0-1-0-1… 1 0 1 0 1 0 3 127 Cxcl9
## 4 1-0-1-0-1… 1 0 1 0 1 0 3 127 Acod1
## 5 1-0-1-0-1… 1 0 1 0 1 0 3 127 Cxcl10
## 6 1-0-1-0-1… 1 0 1 0 1 0 3 127 Socs3
## 7 1-0-1-0-1… 1 0 1 0 1 0 3 127 Olr1
## 8 1-0-1-0-1… 1 0 1 0 1 0 3 127 Gm121…
## 9 1-0-1-0-1… 1 0 1 0 1 0 3 127 Cxcl13
## 10 1-0-1-0-1… 1 0 1 0 1 0 3 127 Gm122…
## # … with 4,423 more rowsWrite into a file 'Grouped_genes.txt'
df_grouped_genes <- tibble(code=df_full %>% pull(code) %>% unique() %>% sort(), group=c('LTr','LTi','MTr','MPr','MTi','MPi','EPi')) %>% mutate(group=str_c(code,' (',group,')')) %>% inner_join(df_full, by='code') %>% select(-code,-ncode,-freq)
df_grouped_genes %>% write_delim('Grouped_genes.txt',delim='\t')